- Journal List
- NIHPA Author Manuscripts
- PMC3179915
- Performing your original search, Scalable and interactive segmentation and visualization of neural processes in EM datasets., in PMC will retrieve 17 records.
Scalable and Interactive Segmentation and Visualization of Neural Processes in EM Datasets
Abstract
Recent advances in scanning technology provide high resolution EM (Electron Microscopy) datasets that allow neuroscientists to reconstruct complex neural connections in a nervous system. However, due to the enormous size and complexity of the resulting data, segmentation and visualization of neural processes in EM data is usually a difficult and very time-consuming task. In this paper, we present NeuroTrace, a novel EM volume segmentation and visualization system that consists of two parts: a semi-automatic multiphase level set segmentation with 3D tracking for reconstruction of neural processes, and a specialized volume rendering approach for visualization of EM volumes. It employs view-dependent on-demand filtering and evaluation of a local histogram edge metric, as well as on-the-fly interpolation and ray-casting of implicit surfaces for segmented neural structures. Both methods are implemented on the GPU for interactive performance. NeuroTrace is designed to be scalable to large datasets and data-parallel hardware architectures. A comparison of NeuroTrace with a commonly used manual EM segmentation tool shows that our interactive workflow is faster and easier to use for the reconstruction of complex neural processes.
1 Introduction
The reconstruction of neural connections to understand the function of the brain is an emerging and active research area in bioscience that is often called Connectomics [28]. With the advent of high-resolution scanning technologies such as 3D light-microscopy and electron microscopy (EM), reconstruction of complex 3D neural circuits from large volumes of neural tissues has become feasible. Among them, however, only EM data can provide sufficient resolution to identify synapses and to resolve extremely narrow neural processes such as dendritic spines of roughly 50 nm in diameter. Current EM technologies are able to attain resolutions of 3–5 nanometers per pixel in the x–y plane. Due to its extremely high resolution, an EM scan of a single section from a small tissue sample can easily be as large as tens of gigabytes, and the total scan of a tissue sample as large as several terabytes of raw data.
These high-resolution, large-scale datasets are crucial for reconstruction of detailed neural connections, but pose very challenging problems for 3D segmentation and visualization. First, the current common practice for segmentation of objects of interest in EM datasets is a mostly manual process, which is very labor-intensive and time-consuming. Even though there have been research efforts to develop automated EM segmentation algorithms, they are not robust enough to deal with common artifacts of real datasets, such as noise and misalignment. Second, the complex structure of nerve cells makes direct volume rendering of EM datasets very difficult. Transfer functions based solely on image intensity and gradient result in cluttered renderings, which degrades visualization quality. Finally, it is important that the segmentation and visualization algorithms are scalable, to cope with the ever-increasing data sizes, while maintaining interactive performance, so that the user can perform manual modifications at any time if necessary.
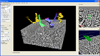
In this paper we present NeuroTrace, a system for segmentation of neural processes in high-resolution EM data that integrates semi-automatic segmentation and centerline tracking with advanced volume visualization. The resulting workflow improves the current state-of-the-art approach of neurobiologists significantly. Our first contribution is a novel interactive 3D segmentation approach that is based on a sequence of 2D segmentations of cell membranes using active ribbons [32]. By integrating an image correspondence energy into the level set formulation we achieve robust transition between consecutive slices. Using these 2D segmentations and a tracking method with weighted path extrapolation we can robustly trace a 3D centerline of a neural pathway along non-axis aligned slices. The second contribution is a volume rendering method with on-demand filtering for de-noising and detection of structure boundaries. A local histogram-based edge metric provides better visual cues to easily find regions of interest in complex EM datasets compared to traditional transfer functions. A third contribution is the efficient implementation of these algorithms on the GPU. We use a dynamic out-of-core caching system to ensure scalability to arbitrary input data sizes. A fourth contribution is a combined high-quality visualization of the volume and the segmented neural processes, combining direct volume rendering and implicit surface ray-casting in a single rendering pass. Our final contribution is an integrated workflow that provides a unified user-interface to easily explore large EM volumes and extract neural processes at interactive rates (Figure 1). The results of our user study show that NeuroTrace is more efficient and accurate than the leading segmentation tool.
2 Previous Work
Automated EM Segmentation
Large scale EM reconstruction with automated methods has only very recently gained much attention. Jurrus et al. [13] proposed an automated method to trace axons in serial block-face scanning EM datasets. Their method uses iterative Kalman filtering together with an active contour model and a vector field produced by an optical flow method to estimate the axon location on each slice. Macke et al. [17] proposed a probabilistic framework to guide level set propagation on each slice, where the probabilistic framework models the similarity between slices. Mishchenko [21] proposed a 3D neural reconstruction method consisting of a Hessian-based 2D ridge detector to extract axon boundaries and a weighted graph clustering method to generate a connectivity map across slices. All these methods assume that the neural processes follow a specific direction, usually orthogonal to the scanning plane. However, this assumption fails for many axons. The non-axis aligned arbitrary 3D tracking method proposed in this paper provides much more flexibility to handle axons at various orientations. Bartesaghi et al. [2] use 3D minimal surface to segment cell boundaries in high resolution electron tomograms. However, their method cannot be directly applied to segment elongated structures as in our case because the method assumes the target structure is spherical topology.
Vessel Extraction and Virtual Endoscopy
Segmentation and tracking of thin structures is an ongoing area of medical research. Kirbas et al. [15] give an extensive review of automated vessel extraction methods. In virtual endoscopy, the focus is on 3D path planning inside elongated structures (e.g., [11, 3, 7]). However, these approaches either require already segmented data or are not directly applicable to high-resolution EM volumes because of the more complicated structures involved.
Volume Rendering of EM Data
Volume rendering of microscopic structures is a very recent area of research. Mayerich et al. [20] segment and visualize microvascular structures and their relationships, but the resolution is two orders of magnitude lower than EM data. We employ GPU-based ray-casting of volumes and implicit surfaces [25] using a bricking scheme for large data [4] implemented in CUDA. Enhancing edges or structure boundaries has always been important in volume rendering, and is typically achieved using higher-order transfer functions [14]. Caban et al. [5] have recently introduced texture-based tranfer functions based on first-, second-, and high-order local (histogram) statistics. However, these methods are not effective in dealing with noise in EM images. Our rendering framework employs a general filtering and de-noising step with a neighborhood size that can be changed interactively. Martin et al. [19] define a set of brightness, color, and texture cues for constructing a local boundary model. To enhance edges during ray-casting we extended their 2D boundary detection framework using local histogram comparisons.
Iso-Surface Rendering
Rendering elongated structures with elliptical cross-sections has been of interest in diffusion tensor imaging (DTI). Interpolation between successive ellipses has been used together with ray-casting [24], as well as with a geometry setup stage [23]. Instead of targeting many relatively thin fibres, we use interpolating quaternion frames between successive ellipses. Our method requires few evaluations of trigonometric functions, which enables high rendering performance and simple implementation.
3 Workflow
We assume that registration of the EM data is performed in a pre-processing step. The individual image tiles acquired by the cameras in the EM are warped and stitched together to obtain slice images of very high resolution. These slices are then registered in 3D so that they are aligned and structures can be followed from slice to slice.
3.1 Current Practice
There are a number of manual tools for the segmentations of EM datasets [27]. The software package most commonly used by neuroscientists is Reconstruct [9]. The main window of this tool displays a 2D axis-aligned view of the current slice. To identify the structures of interest, users can move from one slice to the next and inspect each in turn using basic viewing functions such as zoom, pan, scale, and rotation. The segmentation is manual, using polygon, curve, and free-form drawing tools. The final segmented neural processes can be rendered as 3D polygon meshes. To generate higher-quality images, the scientists often use additional volume visualization packages, such as Amira (http://www.amiravis.com).
This workflow is straightforward but also very labor-intensive and time-consuming. It lacks integrated volume visualization of the input data and high-quality visualization of the resulting segmentation. Reconstruct only allows axis-aligned tracking, so neural processes parallel to the image are difficult to segment. In addition, the data has to fit into main memory, which limits the scalability of the system.
3.2 Proposed Workflow
Figure 2 illustrates our integrated, interactive workflow for visualizing and segmenting neural processes. The first step in our workflow is to inspect the input volume using volume rendering before any segmentation is performed in order to obtain an overview and to determine a region of interest (ROI) (Figure 1 middle). In order to better delineate the structures of interest we modified the volume rendering such that the boundaries of neural processes are depicted more clearly (Section 5).

Using the 3D volume view, the user can specify the center of the current ROI on an arbitrarily oriented 2D clipping plane. The corresponding oblique slice is then shown in an additional 2D view (Figure 1 top right). The next step is to quickly paint a rough approximation of a boundary of interest, e.g., of an axon, in this 2D view. This input is used to initialize an active ribbon that automatically performs tracking of the cell boundary from slice to slice (Section 4). The individual cell boundaries are shown in the 2D view and can be inspected and modified interactively at any time.
While the segmentation and tracking is in progress, the segmentation obtained thus far is concurrently shown in the 3D volume view (Figure 1 middle). The long, elongated structures that are of highest importance in our application can be represented well by elliptical cross-sections. Therefore, in order to use as little memory as possible, an ellipse is fitted to the active ribbon in each slice. These ellipses are interpolated on-the-fly during volume ray-casting in order to obtain smooth, connected implicit surfaces in 3D (Section 5.2). This process is repeated for every neural process of interest, iteratively adding additional structures, which are all depicted concurrently in 3D.
The main advantage of our workflow is that it tightly integrates semi-automatic segmentation and visualization, which allows the user to inspect and modify the ongoing tracking and segmentation process at any time, while minimizing the amount of user interaction that is necessary.
4 Segmentation
We compute 3D segmentations using a combination of 2D neural membrane segmentations and 3D centerline tracking. For 2D segmentation we use a modified multiphase level set active ribbon model originally proposed by Vazquez et al. [32] (Figure 3). Because level set segmentation is very sensitive to initialization, we propose a novel active ribbon formulation by adding an additional constraint based on image correspondence between current and previous slices. Once 2D segmentation is done, we extrapolate the next point along the centerline of the possibly non-axis aligned neural process.

4.1 2D Neural Membrane Segmentation
The active ribbon model is based on two deformable moving interfaces (ϕ1 and ϕ2 in Figure 3 left) interacting with each other to maintain ribbon topology. The level set equation for each ϕi of the active ribbon model is defined as follows:
where
is the data dependent speed to move toward the membrane boundary, c1 and c2 are the average pixel intensity of inner cell region and cell membrane, respectively, and I is the pixel intensity of the input image.
is the ribbon consistency speed to keep constant distance between two level set interfaces ϕi and ϕj. σi (ϕj) returns a positive value if two level sets are too close and a negative value otherwise (more details can be found in [32]). FK is the mean curvature speed to maintain the smoothness of the interfaces. Because two interfaces push or pull each other until they converge to the target, the active ribbon model is very robust in noisy and feature-rich EM images. In addition, the model includes a force field that allows neighboring ribbons to interact with each other.
However, Vazques et al. [32] assume that neural processes are orthogonal to the image plane and that there is no large displacement between consecutive slices, which is typically not the case. In addition, the cross sectional shape of the neural process may deform significantly between slices, making initialization of the active ribbon challenging. Therefore, we propose a new active ribbon formulation by adding a force field that maps one image to another using image correspondence. This allows us to robustly initialize the location of the neural membranes on subsequent slices.
Let Ii and Ii+1 be two consecutive slices, where each slice is defined on a 2D domain Ω. We can define the energy between two images for a given vector field v, which describes how different two images are, as follows:
where Ĩi is the image Ii deformed by the vector field v, and α is a regularization parameter. Finding the vector field v that minimizes Equation 2 is a nonrigid image registration problem [1, 6]. In order to find v, we use a gradient flow method along the negative gradient direction of EI with respect to v. To avoid local minima and to handle large deformations more efficiently, we use a multilevel approach and compute the solution on different scales, from coarse to fine.
Once we have the vector field v, we can define the energy Eϕ that measures the difference between two distance fields ϕi and ϕi+1 for the images Ii and Ii+1 as follows:
where ![[phi with tilde]](./Scalable and Interactive Segmentation and Visualization of Neural Processes in EM Datasets_files/x03D5x0303.gif) i is ϕi deformed by v. Note that ϕi and ϕi+1 in this discussion are not the inner and outer level set for the active ribbon (Figure 3 left) but a single level set on two different images. Thus we can define the level set function for ϕi+1 that minimizes Eϕ as follows:
i is ϕi deformed by v. Note that ϕi and ϕi+1 in this discussion are not the inner and outer level set for the active ribbon (Figure 3 left) but a single level set on two different images. Thus we can define the level set function for ϕi+1 that minimizes Eϕ as follows:
where FC is the image correspondence speed and γ is a level set parameter. The image correspondence speed FC can be defined using the gradient of Eϕ with respect to ϕ as follows:
The image correspondence speed FC can be integrated into the level set equation 1 like other speed functions. In our implementation, we gradually decrease ξ as the level set iteration proceeds so that the entire active ribbon can move towards the correct location of the target membrane at the beginning, and then becomes more stable at the end such that the ribbon boundaries can close in on the membrane boundaries. Figure 4 shows the robust transition of the active ribbon between slices with the image correspondence force.
4.2 3D Centerline Tracking
To deal with non-axis aligned neural processes, we implemented a tracking algorithm that follows the centerline of the process. Even though tracking a centerline through membrane centers may seem straightforward, it is not simple in our case because we do not know membrane locations in advance. In other words, even though the current slice position and segmentation are given, we do not know the position and segmentation of the next slice.
To tackle this problem, we propose a two-step method that consists of estimation and correction steps. In the estimation step, the tangent direction Vt at the last center point is computed using a one-sided finite difference method. We also keep the previous tracking direction Vp. The new tracking direction is then the weighted average between those two vectors: V = ωVp + (1 − ω)Vt (Figure 3 right). The weight ω controls the amount of history used to determine the current tracking direction. We typically use a value of ω = 0.9 for smooth transition between slices.
Once we compute a new tracking direction, a temporary new center position Ci+1 of the next slice can be estimated by simple extrapolation as 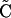 i+1 = Ci + δV, where δ is the pixel width (i.e., grid spacing) in order to move no more than one pixel distance per estimation step. The local frame of the previous slice is then projected onto the new plane defined by the center
i+1 = Ci + δV, where δ is the pixel width (i.e., grid spacing) in order to move no more than one pixel distance per estimation step. The local frame of the previous slice is then projected onto the new plane defined by the center 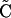 i+1 and the normal V. A new 2D slice is resampled from the volume data using the new local frame and used for segmentation. Finally, in the correction step,
i+1 and the normal V. A new 2D slice is resampled from the volume data using the new local frame and used for segmentation. Finally, in the correction step, 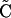 i+1 is replaced by the correct center of the segmented neural membrane, Ci+1. Figure 5 shows an example of 3D centerline tracking and segmentation using NeuroTrace.
i+1 is replaced by the correct center of the segmented neural membrane, Ci+1. Figure 5 shows an example of 3D centerline tracking and segmentation using NeuroTrace.
4.3 GPU Implementation
Our GPU level set solver updates the level set only in active regions using a block-based narrow band proposed by Lefohn et al. [16]. A slight difference is that we collect all the blocks within a user-defined narrow band size, where the minimum distance to the zero level set of each block is computed in the redistance step without explicitly checking the activation of neighboring blocks. The main level set update process consists of four steps: (1) Form the active list by collecting the active blocks. (2) Iteratively update the level set on each active block in the active list up to the pre-computed number of iterations (based on the narrowband width). (3) Recompute the distance from the zero level set. (4) Stop if the level set converges to a steady state or the maximum number of iterations is reached. Otherwise go to (1).
The active list is a one dimensional array of unsigned integers. The first element in this list is the total number of active blocks, and the rest of the array contains the active block indices. To manage the active list efficiently, we store it entirely on the GPU. The only interaction between the CPU and the GPU is copying the first element of the active list from the GPU to the CPU. Then the host code launches a CUDA kernel with the grid size equal to the total number of active blocks. The size of a CUDA block is the same as an active block for our level set. In the CUDA kernel, the global memory address is computed by off-setting from the base address using the active block index. Managing the active list, i.e., adding new active blocks and removing non-active blocks, can be achieved using the atomic hardware operators of recent NVIDIA GPUs without using additional stream compaction processes. We can compute the minimum distance to the zero level set for each block using parallel reduction. If the minimum distance is smaller than the user-defined narrow band width, the total number of active blocks is increased by one using AtomicAdd (). Then the current block index is stored at the end of the current list using the index returned by the atomic operator.
Once the active list is formed, then each block in the list can be up-dated multiple times depending on the width of the narrow band. For example, if the grid spacing is 1 and the width of the narrow band is 10, then we can safely update the active blocks in the current active list 10 times without refreshing the active list (i.e., explicitly checking the (de-)activation of the blocks). This is because the CourantFriedrichsLewy (CFL) condition [26] guarantees that the maximum deformation incurred by a single update of the level set cannot be greater than the grid spacing. The level set update is done using a Jacobi update method, and communication between block boundaries can be handled implicitly by calling the new CUDA kernel for each level set update because the new solutions are written back to global memory after each update.
In extending the single level set method to multiphase level sets we need to evaluate the correct distance between two level sets to keep the topology of the active ribbon consistent. However, the active ribbon does not guarantee the correct distance after deformation due to the combination of various force fields. Therefore, we recompute the distance field for each level set when the list of active blocks is up-dated. Note that we need to redistance not only on the active lists but the complete level sets because the level sets may not share the same active list unless they are very close to each other. To quickly compute the distance fields we employ the GPU-based Eikonal solver by Jeong etal. [12].
We implemented the nonrigid image registration method using semi-implicit discretization as a two-step iterative process, updating and smoothing the vector field v as follows:
where G is a Gaussian smoothing kernel. Equation 6 is a simple Euler integration that can be efficiently mapped to the GPU. To interpolate the pixel values Ĩi and ![[nabla]](./Scalable and Interactive Segmentation and Visualization of Neural Processes in EM Datasets_files/nabla.gif) Ĩi on locations defined by v we use texture hardware interpolation on the GPU. Texture memory is cached, so it is efficient for locally coherent random memory accesses. To speed up the 2D Gaussian smoothing in image space, we implemented a sequence of 1D convolutions using shared memory and apply them along x and y, respectively.
Ĩi on locations defined by v we use texture hardware interpolation on the GPU. Texture memory is cached, so it is efficient for locally coherent random memory accesses. To speed up the 2D Gaussian smoothing in image space, we implemented a sequence of 1D convolutions using shared memory and apply them along x and y, respectively.
5 Volume Visualization
Volume rendering of high-resolution EM data poses several challenges. EM data is extremely dense and heavily textured, exhibits a complex structure of interconnected nerve cells, and has a low signal-to-noise ratio. Therefore, standard volume rendering results in cluttered images that make it hard to identify regions of interest (ROIs) or to observe an ongoing segmentation.
Our visualization approach supports the inspection of data prior to segmentation, for identifying ROIs, as well as the visualization of the ongoing and final segmentation (see Figure 2). To improve the visualization of the raw data prior to segmentation, we have implemented on-the-fly nonlinear noise removal and edge enhancement to support the user in finding and selecting ROIs. Using a local histogram-based edge metric, which is only calculated on demand for currently visible parts of the volume and cached for later reuse, we can enhance important structures (e.g., myelinated axons) while fading out less important regions. During ray-casting we use the computed edge values to modulate the current sample’s opacity with different user-selectable opacity weighting modes (e.g., min, max, alpha blending).
5.1 On-demand Filtering
The main motivations for on-demand filtering (i.e., noise removal and edge detection) are the flexibility offered by being able to change filters and filter parameters on the fly while avoiding additional disk storage and bandwidth bottlenecks for terabyte-sized volume data. We perform filtering only on blocks of the volume that are visible from the current viewpoint, and store the computed data directly on the GPU for later reuse. We have implemented a caching scheme for these pre-computed blocks on the GPU to avoid costly transfers to and from GPU memory while at the same time avoiding repetitive recalculation of filtered blocks. During visualization we display either the original volume, the noise-reduced data, the computed edge values, or a combination of the above.
Our on-demand filtering algorithm consists of several steps: (1) Detect for each block in the volume if it is visible from the current viewpoint. (2) Build the list of blocks that need to be computed. (3) Perform noise removal filtering on selected blocks and store them in the cache. (4) Calculate the histogram-based edge metric on selected blocks and store those blocks in the cache. (5) High-resolution ray-casting combining edge values and original data values. The detection of visible blocks (Step 1) is done either in a separate low-resolution ray-casting pass or included in Step 5.
5.1.1 Noise Removal
Since EM data generally exhibits a low signal-to-noise ratio we have integrated an on-demand noise removal filter step into our pipeline prior to calculating the local histogram-based edge metric. We perform the filtering only on those blocks that were marked as visible and are not present in the cache yet. We have implemented 2D and 3D Gaussian, mean, non-linear median, bilateral [31], and anisotropic diffusion filters [22] with user adjustable neighborhood sizes. Especially non-linear filters have shown good noise removal properties without degrading edges in the EM data [30]. Our main objective, however, was to develop a general framework for noise removal, where additional filters could be added easily. The results for each processed block is stored in the cache and used as input for the edge detection algorithm.
5.1.2 Local Histogram-based Edge Detection
We use a local histogram-based edge metric to modulate the opacity of the EM data during raycasting. Boundaries in the volume get enhanced while more homogenous regions are supressed. This helps the user in navigating through the unsegmented dataset and in finding regions where a segmentation should be started. The edge metric is computed only for visible blocks that are not stored in the cache yet.
Our edge detection algorithm is based on the work of Martin et al. [19] who introduced edge and boundary detection in 2D image based on local histograms. They did a thorough evaluation of different brightness, color, and texture cues for constructing a local boundary model, which was subsequently used to detect contours [18] in natural images.
In our local histogram-based edge detection approach we take a block neighborhood around each voxel to calculate the brightness gradient for different directions. We separate the voxel’s neighborhood along the given direction into two halves and calculate the histogram in each half-space. Finally, the histogram difference is calculated using the χ2 distance metric. A high difference between histograms indicates an abrupt change in brightness in the volume, i.e., an edge. The maximum difference value over all directions is saved as the edge value in the cache block. As the neighborhood size for the histogram calculation can be adjusted to match the resolution level of the current input data, this approach scales to large data and to volume subdivision schemes like octrees. Again, we have kept the implementation of our edge detection framework as modular as possible to support adding different edge detection algorithms in the future. During volume rendering, we fetch at each sample location the corresponding edge value and use it to modulate the sample’s opacity and/or color. Optionally, the user can first use a windowing function on the calculated edge values to further enhance the visualization.
5.1.3 Dynamic Caching
To improve the performance of our edge-based visualization scheme we have implemented a dynamic caching scheme for storing on-the-fly computed blocks. Two caches are allocated directly on the GPU, one to store de-noised volume blocks and the second to store blocks containing the calculated edge values. First, the visibility of all blocks is updated for the current viewpoint in a first ray-casting pass and saved in a 3D array corresponding to the number of blocks in the volume. Next, all blocks are flagged as either: (1) visible, present in cache; (2) visible, not present in cache; (3) not visible, present in cache; or (4) not visible, not present in cache. Visible blocks that are already in the cache (flagged with (1)) do not need to be recomputed. Only blocks flagged with (2) need to the processed. Therefore, indices of blocks flagged with (2) are stored for later calculation (see Section 5.1.4). During filtering/edge detection the computed blocks are stored in the corresponding cache. A small lookup table is maintained for mapping between block storage space in the cache to actual volume blocks as described in [4]. Unused blocks are kept in the cache for later reuse (flagged with (3)). However, if cache memory gets low, unused blocks are flushed from the cache and replaced by currently visible blocks.
5.1.4 GPU Implementation
After detecting which blocks need processing, a CUDA kernel is launched with grid size corresponding to the number of blocks that need to be processed. For simplicity we explain the implementation of our filtering and edge detection algorithm in 2D. The extension to 3D is straightforward.
To calculate filter/edge values in each block, we start a CUDA kernel with a CUDA block size that corresponds to the user specified neighborhood size, but with one dimension less than the actual neighborhood (e.g., for a 3D neighborhood a 2D CUDA block is started, for a 2D neighborhood a ID CUDA block is started). Figure 6 depicts the case where the edge detection of a block uses a 5×5 neighborhood. In this case the kernel is started with 5 concurrent threads. Next, the threads iterate over the entire block that needs to be filtered and calculate the filter/edge values for each voxel. Each thread is responsible for only one part of the filter’s neighborhood, as depicted by the colored areas in Figure 6. To reduce redundant texture fetches each thread locally caches its last computed values. The size of this thread-local array corresponds to the neighborhood size of the filter. Therefore, at each step a thread only needs to perform one texture fetch, and store the value in its local cache (Figure 6, middle).

To calculate the local-histogram based edge metric, all samples in a voxel’s neighborhood need to be assigned to one of the two local histograms (for both half-spaces), as depicted in Figure 6, right. The histograms are stored in shared CUDA memory and used for the final calculation of the χ2 histogram difference. The main steps for each thread are: (1) Update the histogram of the first half-space (histogramleft) by removing the sample that has left the filter neighborhood and adding the last sample from histogramright. (2) Remove the sample that has left the filter neighborhood from the thread-local cache. (3) Fetch the sample that has entered the filter neighborhood from the volume texture and store it in the thread-local cache. (4) Update the histogram of the second half-space (histogramright) by removing the sample that is now in histogramleft and adding the sample that has just been fetched from the volume texture. All threads are synchronized after they have performed the above steps using atomic CUDA operations for updating the shared histograms. Now the χ2 histogram difference for the current neighborhood can be computed and stored in the cache.
To implement the de-noising filters we use the same basic strategy. For Gaussian filters we transfer a 1D look-up table of the weights to the GPU to speed up the calculation. For bilateral filtering we use the same look-up table to calculate the geometric closeness function, whereas the photometric similarity function is calculated on-the-fly in the CUDA kernel. For median filtering we implemented bitonic sort on the GPU to find the median value of the filter neighborhood. Anisotropic diffusion filtering is the most complex filter in our framework. It requires a second filter cache to allow ping-pong swaps between source and destination. Also, costly neighborhood lookups in the source cache are needed to compute the boundary values of the destination blocks.
If the noise removal step is performed prior to the edge detection, the local histogram calculation uses the values from the filtered block cache as input values instead of the original volume texture. Therefore, special care has to be taken when fetching de-noised values for a neighborhood at an edge-block’s boundary. This case can be handled by either extending the dimensions of the de-noised blocks compared to the edge-detected blocks, or by detecting which additional blocks would have to be de-noised and performing a neighborhood lookup for areas outside an edge block’ boundary.
5.2 Visualization of Segmented Neural Structures
In order to visualize and inspect the segmented neural processes in 3D, we depict the original volume data together with semi-transparent iso-surfaces that delineate structures such as axons or dendrites. The output image is generated in a single ray-casting pass for both the iso-surfaces and the part of the volume that is shown using direct volume rendering. While stepping from sample to sample along a given viewing ray, the direct volume rendering integral is solved via front-to-back compositing. At the same time, each sample is tested for potential intersections with iso-surfaces. If a surface is intersected, its color and transparency are composited with the accumulated volume-rendered part, and direct volume rendering is continued behind the surface intersection.
Our active ribbon segmentation described in Section 4 outputs a set of implicit surfaces for each 2D slice. However, in order to make the system scalable for large EM data, we do not store these 2D distance fields. Instead, we convert the segmentation to a very compact format by fitting an ellipse to each active ribbon. An entire structure such as an axon is then represented as a simple list of elliptical cross-sections, which reduces the memory footprint significantly.
5.2.1 Implicit Surfaces from Elliptical Cross-Sections
To render smooth, connected surfaces from elliptical cross-sections we compute implicit surfaces from the set of ellipses on-the-fly. Although everything is computed in a single ray-casting pass, in order to simplify the problem we treat it as two conceptually separate parts. The first part is ray-casting of an implicit surface in a distance field ϕ(x), where the surface is defined by the points where ϕ(x) = 0. The second part is the computation of ϕ(x) for any point x in volume space. Here, a point x is either a sample p on a viewing ray, or the location of a central differences computation during shading.
The ray-caster renders and shades implicit surfaces by evaluating ϕ(p) along viewing rays. Intersection with an implicit surface is detected between two successive points on a ray when ϕ(pi) < 0 and ϕ(pi+1) > 0, i.e., the first sample is in front and the second one behind the surface. We carry out a predetermined number of bisection steps in order to find a sufficiently accurate intersection position [10]. At that location, we compute the shading using the normalized gradient obtained from central differences in the distance field, evaluating ϕ(x) at six additional locations. In order to simplify accommodating multiple axons that are represented as implicit surfaces each, we map every point x in volume space for surface intersection and shading purposes to only one distance ϕ(x), which is the distance to the closest axon. Conceptually, each axon is represented by a 3D distance field ϕi, and the ϕ(x) used in the ray-casting loop is ϕ(x) = mini (ϕi(x)).
In order to obtain implicit surfaces in 3D from a collection of 2D elliptical cross-sections, we have to be able to evaluate ϕ(x) throughout the volume. We want this interpolation to be fast and easy to implement, and still result in smooth shaded surfaces. Each cross-section is represented by a single ellipse elli = (ci,qi,
The main idea for evaluating ϕ(p) at a given point p is to directly compute a single interpolated ellipse whose plane exactly or almost contains p, and then to perform a straight-forward 2D point-to-ellipse distance computation in this plane. That is, we first compute an interpolated ellipse ell(p) = (cp,qp, ![[set membership]](./Scalable and Interactive Segmentation and Visualization of Neural Processes in EM Datasets_files/x2208.gif) [0, 1] is required for a given p, which we compute as follows:
[0, 1] is required for a given p, which we compute as follows:
This is illustrated in Figure 7 (left).
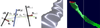
We compute α such that it is always 0 in the plane of elli, and 1 in the plane of elli+1, which guarantees that successive segments between ellipse pairs line up exactly. We require a vector ![[n with macron]](./Scalable and Interactive Segmentation and Visualization of Neural Processes in EM Datasets_files/x006Ex0304.gif) that is guaranteed not to be parallel to either ellipse, and compute α as the ratio of k0, the distance from p along
that is guaranteed not to be parallel to either ellipse, and compute α as the ratio of k0, the distance from p along ![[n with macron]](./Scalable and Interactive Segmentation and Visualization of Neural Processes in EM Datasets_files/x006Ex0304.gif) to elli, to k0 + k1, the total distance between elli and elli+1 along ñ through p. We have chosen ñ as the half-way vector between ni and ni+1. This choice fulfills our requirements and yields smooth results. Another obvious choice would be ci+1 − ci. However, in our case this vector can be close to parallel to the ni, which can result in numerical problems in the denominators of k0 and k1 (Equation 8).
to elli, to k0 + k1, the total distance between elli and elli+1 along ñ through p. We have chosen ñ as the half-way vector between ni and ni+1. This choice fulfills our requirements and yields smooth results. Another obvious choice would be ci+1 − ci. However, in our case this vector can be close to parallel to the ni, which can result in numerical problems in the denominators of k0 and k1 (Equation 8).
After α has been computed, it is used to obtain ell(p) as linear interpolation between the ellipse centers and axis lengths, yielding cp,
This approach gives completely accurate results for parallel ellipse planes, which is a common case in axon tracking where the planes are often orthogonal to the z axis. It is an approximate solution for non-parallel planes that works well in practice. The angle between two successive ellipse planes ni and ni+1 is always quite small, even though the whole axon is allowed to curve significantly from the first cross-section to the last. Figure 7 (middle) shows a close-up of an axon with non-parallel ellipse planes, which illustrates that our approach results in visually smooth results.
5.2.2 GPU Implementation
In order to speed up finding the two ellipses nearest to a given point p in the CUDA ray-casting kernel, ellipses are sorted into a 3D block structure (e.g., 163 blocks) before rendering that only needs to be up-dated when new ellipses are added. Each block contains links (integer indices) to all ellipses intersecting it. A single ellipse can be linked to by several blocks, but during rendering only a single block needs to be examined for each point p. In order to efficiently handle empty blocks, each block only stores the number of ellipses that intersect it and a start index into a global array of links to ellipses. The array is packed tightly such that all links of non-empty blocks are stored at consecutive memory locations. Actual ellipse information (axon-ID,ci,qi,
6 Results
We implemented our segmentation and visualizations system on a Windows XP PC equipped with quad-core Intel Xeon 3.0 GHz CPU, 16 Gigabytes main memory, and NVIDIA Quadro 5800 and Tesla C1060 GPUs. We used a single CPU core and one GPU to compare the running time on each architecture. The CPU version is implemented using the ITK image processing library (http://www.itk.org). The main computational code is similar on the CPU and GPU for a fair comparison.
6.1 Segmentation
The running time of the CPU level set solver for 100 iterations on a 512 × 512 image is 7 seconds. It is only 0.3 second on the GPU, which shows about 23 times speed-up. Our GPU image registration runs less than a second on a 512 × 512 image (500 iterations). The total running time of our segmentation method per slice, without user interaction, is only about a second, which is sufficient for interactive applications.
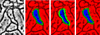
To assess the performance of our segmentation method, we have segmented multiple axons in two EM datasets and measured the total and per-slice times, the amount of user intervention, and the ellipse approximation errors. The first dataset is an adult mouse cortex that consists of 101 slices of 1008 × 1065 2D image, where each pixel has five nanometers resolution and the section thickness is about 30 nanometers. The second dataset is an adult mouse hippocampus that consists of 50 slices of 1278 × 756 2D image, where each pixel is four nanometers wide and the section thickness is 29.4 nanometers. Figure 8 shows 3D renderings of the segmented axons and Table 1 lists the segmentation result for each dataset.
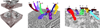
In the mouse cortex dataset, axons A to D were traced using only axis-aligned tracking directions and axons E to H were traced using arbitrary tracking directions. All axons were traced along the z-axis in the mouse hippocampus dataset. Roughly between five to ten percent of the total number of slices were manually edited for correct segmentation for the mouse cortex dataset, and up to 20 percent of the total slices were edited on the mouse hippocampus dataset. Note that the image resolution of our input EM data is up to a factor of five higher than those used in previous work [13, 17]. The data contains more complex neural structures and is very challenging for automated methods. Total times and computing times are not significantly different between axons, and about half of the total time is used for computation.
Our ellipse-based 3D neuron representation can greatly reduce the memory footprint. For example, for an axon of 350 nm diameter we need about 70 × 70 pixels where the pixel width is 5 nm, which requires 9800 floats to store two distance fields. In contrast, to represent an equivalent 3D ellipse we only need to store nine floats, three for center and six for two axis. This yields a compression ratio of more than a factor of a thousand. Table 1 also shows the average distance between the ellipse and the membrane of neurons. The relative ellipse approximation errors, shown in parenthesis, range only between 0.6 to six percent of the longest axis of the ellipse, which is acceptable considering the high compression ratio we achieve.
6.2 Visualization
The prefiltering and edge-detection methods (Figure 9) were both implemented entirely in CUDA and achieve interactive framerates. Filtering blocks on-demand and caching them for later reuse allows the user to change filters and filter settings interactively. Especially de-noising prior to calculating the edge metric improved the results considerably. The best results were achieved using anisotropic diffusion filtering. For our local histogram-based edge metric we found a histogram with 64 bins to be sufficient for our data. Also, a simple average-based histogram difference operator showed good results compared to the computationally more complex χ2 distance metric. For our caching scheme we used 83 sized blocks, but this can be adjusted according to the resolution of the data. At the moment our implementation of the cache is based on CUDA arrays, but in the future we would like to use 3D textures to improve tri-linear filter performance during ray casting.

The dimension of EM data is highly anisotropic, with z-slice distances that can be a factor of 10 or more larger than pixel resolution. This poses real problems for volume visualization, since the visible edges from axons are shifted by large amounts between slices. Even though our filtering and edge detection method works better than traditional transfer functions, the results are sometimes still ambiguous and confusing, requiring closer inspection of the 2D slice views to identify the ROI.
6.3 User Study
We have conducted informal user studies of our segmentation method to assess the usability and accuracy of NeuroTrace by comparing it with Reconstruct [9]. We selected six test subjects in total. Two (Expert 1 and 2) are expert neuroscientists, and the other four (Novice 1 to 4) are novices with no previous neural process segmentation experience. We conducted two user studies, where each study required four test subjects (two experts and two novices) to perform segmentation of the same axon (axon E in the mouse cortex dataset and axon A in the mouse hippocampus dataset). We measured the total time and segmentation accuracy for both systems. We also received qualitative feedback from the users.
To measure the segmentation error, we used the Dice metric [8] that is commonly used to quantitatively measure the accuracy of segmentation algorithms [29]. The Dice metric measures similarity between two sets A and B using 2|A ∩ B|/(|A| + |B|), where | · | indicates set size. In our case, A is the ground truth set of pixels, and B is the set of pixels from the segmentation result. Dice values range between 0 and 1, where 1 implies a perfect match. We compute the Dice value for each 2D segmentation by comparing it to ground truth that was obtained by careful manual segmentation. Table 2 and and33 show the total segmentation times and average Dice values, and Figure 10 and and1111 show plots of Dice values for each slice.


For manual segmentation using Reconstruct there is no significant difference between the two groups in terms of the total time, but the results from the novice users are less accurate than those of the expert users. In contrast, the results using NeuroTrace do not show a significant difference between the two groups, and the novice users usually generated slightly less errors (higher Dice values) than the experts (Table 2). That indicates that the semi-automated NeuroTrace is less prone to lead to human errors. In addition, NeuroTrace provides better segmentation results up to three times faster than Reconstruct. Note that Expert 2 is an exception because he spent longer time than usual and performed very accurate segmentations using Reconstruct. It is also interesting to note that the results of Reconstruct become less accurate over time, especially for novice users (Figure 10 Novice 1 and 2). This can be explained by fatigue due to the laborious manual segmentation.
The users have given highly positive feedbacks about the usability and accuracy of NeuroTrace compared to Reconstruct: “A lot easier to use; more efficient; automatic function is nice; trustworthy” (Novice 1). “Less work-demanding and accurate” (Novice 2). “Automatic segmentation was far easier to use and quicker” (Novice 3). “It is a more practical program to use and all of it’s tools are very helpful and useful” (Novice 4). “It proceeds automatically, can tilt the tracing plane” (Expert 1). “Fast, user friendly, easy to correct; visualization of the segmented data” (Expert 2). The suggestions for improvements include the addition of advanced user interface functions such as browsing of neural tracks and editing previous history, and adaptation to different data modalities, e.g., optical fluorescent confocal microcopy. Our neuroscientist collaborators are currently using NeuroTrace in their Connectomics research.
7 Conclusions and Future Work
In this paper we introduced NeuroTrace, a novel interactive segmentation and visualization system for neural processes in EM volumes. The main contributions are a novel semi-automatic segmentation and 3D tracking method, efficient volume rendering with on-the-fly filters and edge detection, a scalable implementation of these methods on the GPU, and a novel workflow that has been shown to be more accurate and efficient than current practice.
In the future we would like to implement a greater variety of filters and edge-detection approaches (e.g., Canny edge detection). Also we plan to automatically adjust pre-defined filter settings and opacity windowing function depending on the resolution of the input data. The biggest challenge are the extremely large z-slice distances in EM datasets. The integration of shape based-interpolation or directional coherence methods into the volume rendering might be a promising direction to solve this problem. We also would like to extend the current segmentation and tracking method to handle merging and branching of neural processes. Simultaneous tracking of multiple neural processes in a GPU cluster system would be another interesting future direction.
Acknowledgments
This work was supported in part by the National Science Foundation under Grant No. PHY-0835713, the Austrian Research Promotion Agency FFG, Vienna Science and Technology Fund WWTF, the Harvard Initiative in Innovative Computing (IIC), the National Institutes of Health under Grant No. P41-RR12553-10 and U54-EB005149, and through generous support from Microsoft Research and NVIDIA. We thank our biology collaborators Prof. Jeff Lichtman and Prof. Clay Reid from the Harvard Center for Brain Science for their time and the use of their data. We also wish to thank Dr. Juan C. Tapia, Dr. Ju Lu, Thomas Zhihao Luo, May Zhang, Bo Wang, and Robert Cole Hurley for participating in the user study.
Contributor Information
Won-Ki Jeong, School of Engineering and Applied Sciences at Harvard University.
Johanna Beyer, VRV is Center for Virtual Reality and Visualization Research, Inc.
Markus Hadwiger, VRV is Center for Virtual Reality and Visualization Research, Inc.
Amelio Vazquez, School of Engineering and Applied Sciences at Harvard University.
Hanspeter Pfister, School of Engineering and Applied Sciences at Harvard University.
Ross T. Whitaker, Scientific Computing and Imaging Institute at the University of Utah.
References
Formats:
- Article |
- PubReader |
- ePub (beta) |
- PDF (5.2M)
- Real-time visualization of large volume datasets on standard PC hardware.[Comput Methods Programs Biomed...]Xie KYang J, Zhu YM, . Comput Methods Programs Biomed. 2008 May; 90(2):117-23. Epub 2008 Feb 19.
- Ssecrett and NeuroTrace: interactive visualization and analysis tools for large-scale neuroscience data sets.[IEEE Comput Graph Appl. 2010]Jeong WKBeyer J, Hadwiger M, Blue R, Law C, Vazquez-Reina A, Reid RC, Lichtman J, Pfister H, . IEEE Comput Graph Appl. 2010 May-Jun; 30(3):58-70.
- Carving: scalable interactive segmentation of neural volume electron microscopy images.[Med Image Comput Comput Assist...]Straehle CNKöthe U, Knott G, Hamprecht FA, . Med Image Comput Comput Assist Interv. 2011; 14(Pt 1):653-60.
- Volume electron microscopy for neuronal circuit reconstruction.[Curr Opin Neurobiol. 2012]Briggman KLBock DD, . Curr Opin Neurobiol. 2012 Feb; 22(1):154-61. Epub 2011 Nov 24.
- Volume visualization: a technical overview with a focus on medical applications.[J Digit Imaging. 2011]Zhang QEagleson R, Peters TM, . J Digit Imaging. 2011 Aug; 24(4):640-64.
- Computational Tissue Volume Reconstruction of a Peripheral Nerve Using High-Resolution Light-Microscopy and Reconstruct[PLoS ONE. ]Gierthmuehlen M, Freiman TM, Haastert-Talini K, Mueller A, Kaminsky J, Stieglitz T, Plachta DT. PLoS ONE. 8(6)e66191
- DP2: Distributed 3D image segmentation using micro-labor workforce[Bioinformatics. 2013]Giuly RJ, Kim KY, Ellisman MH. Bioinformatics. 2013 May 15; 29(10)1359-1360
- Computer Assisted Assembly of Connectomes from Electron Micrographs: Application to Caenorhabditis elegans[PLoS ONE. ]Xu M, Jarrell TA, Wang Y, Cook SJ, Hall DH, Emmons SW. PLoS ONE. 8(1)e54050
- A survey of GPU-based medical image computing techniques[Quantitative Imaging in Medici...]Shi L, Liu W, Zhang H, Xie Y, Wang D. Quantitative Imaging in Medicine and Surgery. 2012 Sep; 2(3)188-206
- Machines that learn to segment images: a crucial technology for connectomics[Current opinion in neurobiolog...]Jain V, Seung HS, Turaga SC. Current opinion in neurobiology. 2010 Oct; 20(5)653-666
- PubMedPubMedPubMed citations for these articles
- TaxonomyTaxonomyRelated taxonomy entry
- Taxonomy TreeTaxonomy Tree
- Scalable and Interactive Segmentation and Visualization of Neural Processes in E...Scalable and Interactive Segmentation and Visualization of Neural Processes in EM DatasetsNIHPA Author Manuscripts. Nov-Dec 2009; 15(6)1505PMC
Your browsing activity is empty.
Activity recording is turned off.
See more...- Review The human connectome: A structural description of the human brain.[PLoS Comput Biol. 2005]Sporns O, Tononi G, Kötter RPLoS Comput Biol. 2005 Sep; 1(4):e42.
- Axon tracking in serial block-face scanning electron microscopy.[Med Image Anal. 2009]Jurrus E, Hardy M, Tasdizen T, Fletcher PT, Koshevoy P, Chien CB, Denk W, Whitaker RMed Image Anal. 2009 Feb; 13(1):180-8.
- Contour-propagation algorithms for semi-automated reconstruction of neural processes.[J Neurosci Methods. 2008]Macke JH, Maack N, Gupta R, Denk W, Schölkopf B, Borst AJ Neurosci Methods. 2008 Jan 30; 167(2):349-57.
- Automation of 3D reconstruction of neural tissue from large volume of conventional serial section transmission electron micrographs.[J Neurosci Methods. 2009]Mishchenko YJ Neurosci Methods. 2009 Jan 30; 176(2):276-89.
- An energy-based three-dimensional segmentation approach for the quantitative interpretation of electron tomograms.[IEEE Trans Image Process. 2005]Bartesaghi A, Sapiro G, Subramaniam SIEEE Trans Image Process. 2005 Sep; 14(9):1314-23.
- Fast extraction of minimal paths in 3D images and applications to virtual endoscopy.[Med Image Anal. 2001]Deschamps T, Cohen LDMed Image Anal. 2001 Dec; 5(4):281-99.
- Visualization of cellular and microvascular relationships.[IEEE Trans Vis Comput Graph. 2008]Mayerich DM, Abbott L, Keyser JIEEE Trans Vis Comput Graph. 2008 Nov-Dec; 14(6):1611-8.
- Texture-based transfer functions for direct volume rendering.[IEEE Trans Vis Comput Graph. 2008]Caban JJ, Rheingans PIEEE Trans Vis Comput Graph. 2008 Nov-Dec; 14(6):1364-71.
- Learning to detect natural image boundaries using local brightness, color, and texture cues.[IEEE Trans Pattern Anal Mach Intell. 2004]Martin DR, Fowlkes CC, Malik JIEEE Trans Pattern Anal Mach Intell. 2004 May; 26(5):530-49.
- Visualizing whole-brain DTI tractography with GPU-based Tuboids and LoD management.[IEEE Trans Vis Comput Graph. 2007]Petrovic V, Fallon J, Kuester FIEEE Trans Vis Comput Graph. 2007 Nov-Dec; 13(6):1488-95.
- Review Circuit reconstruction tools today.[Curr Opin Neurobiol. 2007]Smith SJCurr Opin Neurobiol. 2007 Oct; 17(5):601-8.
- Reconstruct: a free editor for serial section microscopy.[J Microsc. 2005]Fiala JCJ Microsc. 2005 Apr; 218(Pt 1):52-61.
- Learning to detect natural image boundaries using local brightness, color, and texture cues.[IEEE Trans Pattern Anal Mach Intell. 2004]Martin DR, Fowlkes CC, Malik JIEEE Trans Pattern Anal Mach Intell. 2004 May; 26(5):530-49.
- Axon tracking in serial block-face scanning electron microscopy.[Med Image Anal. 2009]Jurrus E, Hardy M, Tasdizen T, Fletcher PT, Koshevoy P, Chien CB, Denk W, Whitaker RMed Image Anal. 2009 Feb; 13(1):180-8.
- Contour-propagation algorithms for semi-automated reconstruction of neural processes.[J Neurosci Methods. 2008]Macke JH, Maack N, Gupta R, Denk W, Schölkopf B, Borst AJ Neurosci Methods. 2008 Jan 30; 167(2):349-57.
- Reconstruct: a free editor for serial section microscopy.[J Microsc. 2005]Fiala JCJ Microsc. 2005 Apr; 218(Pt 1):52-61.
- MRI tissue classification with neighborhood statistics: a nonparametric, entropy-minimizing approach.[Med Image Comput Comput Assist Interv. 2005]Tasdizen T, Awate SP, Whitaker RT, Foster NLMed Image Comput Comput Assist Interv. 2005; 8(Pt 2):517-25.